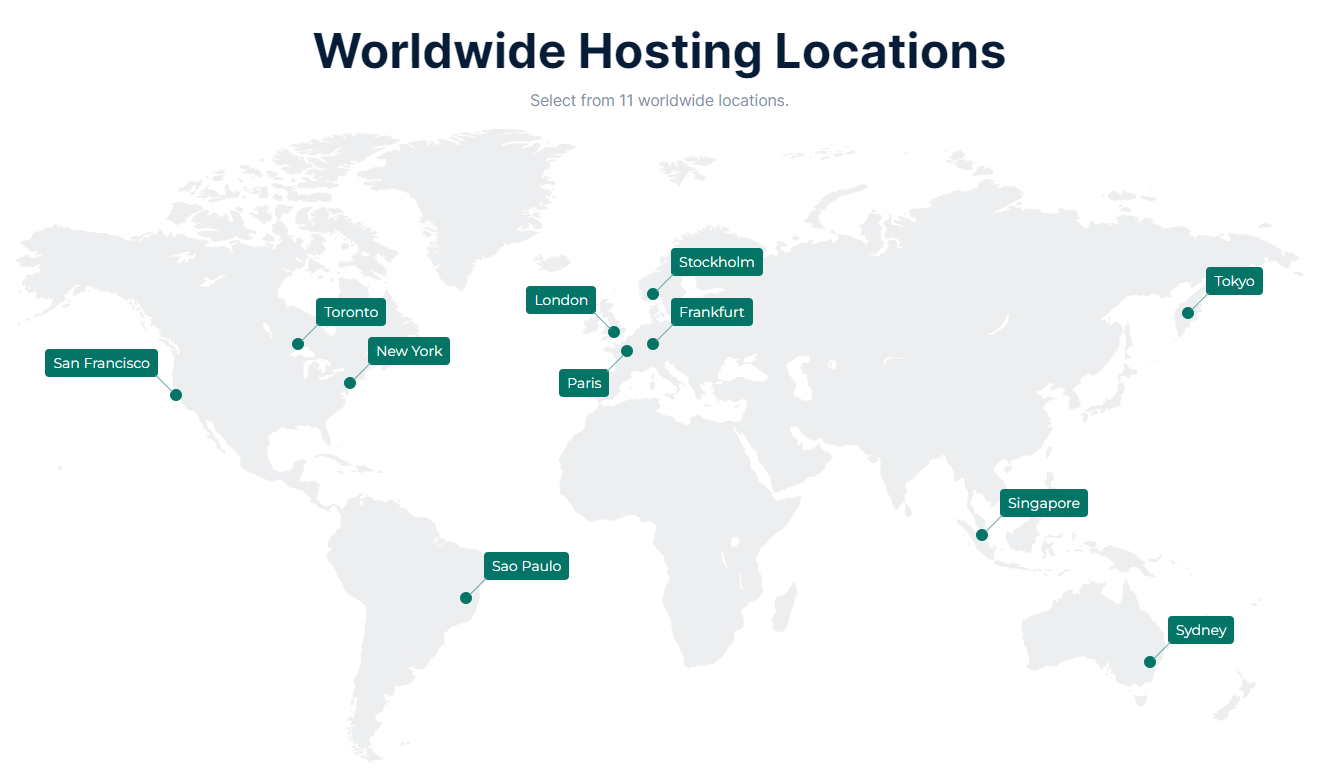
|
|
|
@ -221,8 +221,6 @@ Give your R code a Name and Description.
|
|
|
|
Example
|
|
|
|
Example
|
|
|
|
--------------
|
|
|
|
--------------
|
|
|
|
|
|
|
|
|
|
|
|
The example is animated Plotyl map with Play button.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
The main components in this example are the plotly, ggplot2, RPostgreSQL, and htmlwidgets function.
|
|
|
|
The main components in this example are the plotly, ggplot2, RPostgreSQL, and htmlwidgets function.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
@ -240,8 +238,9 @@ The main components in this example are the plotly, ggplot2, RPostgreSQL, and ht
|
|
|
|
#saveWidget
|
|
|
|
#saveWidget
|
|
|
|
htmlwidgets::saveWidget(as_widget(p), file="index.html")
|
|
|
|
htmlwidgets::saveWidget(as_widget(p), file="index.html")
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
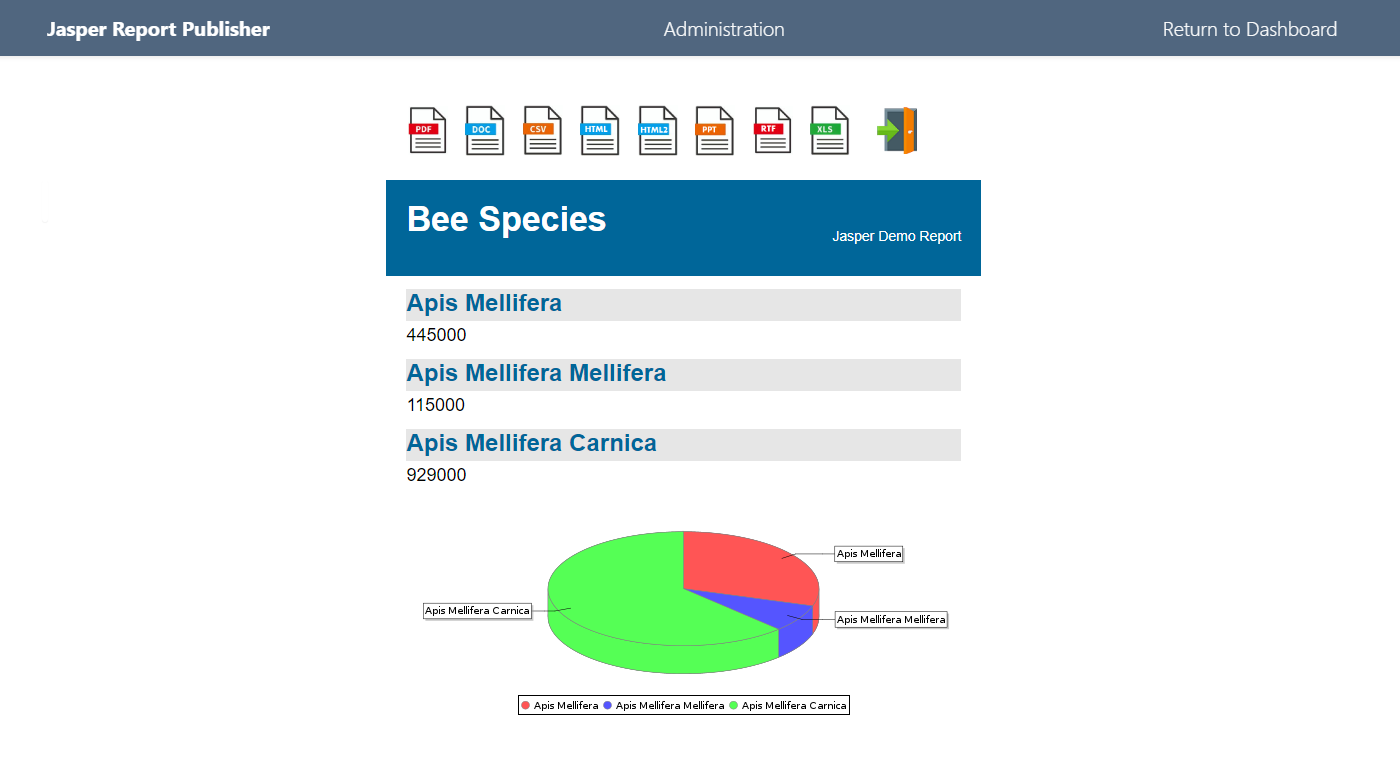
The example is chart with dynamic PostgreSQL connection is contained in the Sample Apps (Simple Bee Harvest)
|
|
|
|
|
|
|
|
|
|
|
|
An example of a Plotly app is included in the installation. Here, we add the RPostgreSQL library to connect to PostgreSQL.
|
|
|
|
Here, we add the RPostgreSQL library to connect to PostgreSQL.
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
.. code-block:: R
|
|
|
|
.. code-block:: R
|
|
|
|
@ -251,27 +250,16 @@ An example of a Plotly app is included in the installation. Here, we add the RP
|
|
|
|
library(RPostgreSQL)
|
|
|
|
library(RPostgreSQL)
|
|
|
|
library(htmlwidgets)
|
|
|
|
library(htmlwidgets)
|
|
|
|
|
|
|
|
|
|
|
|
conn <- RPostgreSQL::dbConnect("PostgreSQL", host = "localhost", dbname = "$DB_NAME", user = "$DB_USER", password = "$DB_PASS")
|
|
|
|
conn <- RPostgreSQL::dbConnect("PostgreSQL", host = "localhost", dbname = "beedatabase", user = "admin1", password = "ORUVDrYBCQ")
|
|
|
|
|
|
|
|
|
|
|
|
query_res <- dbGetQuery(conn, 'SELECT * FROM "sensor_readings";');
|
|
|
|
query_res <- dbGetQuery(conn, 'select area_id,bee_species,sum(average_harvest) from public.apiary GROUP BY (area_id,bee_species) ORDER BY(area_id)');
|
|
|
|
sensor_readings <- as.data.frame(query_res);
|
|
|
|
area_harvest <- as.data.frame(query_res);
|
|
|
|
# sensor_readings$timestamp <- as.Date(sensor_readings$timestamp)
|
|
|
|
|
|
|
|
|
|
|
|
p <- plot_ly(area_harvest, x=~area_id, y=~sum, type="bar",
|
|
|
|
|
|
|
|
text = ~bee_species, textposition = 'auto') %>%
|
|
|
|
|
|
|
|
layout(title = "Accumulated Average Harvest per Area for Apis Mellifera Carnica",
|
|
|
|
|
|
|
|
xaxis = list(title = "Area ID"), yaxis = list(title = "Average Harvest"))
|
|
|
|
|
|
|
|
|
|
|
|
p <- plot_ly(sensor_readings, x=~timestamp, y=~humidity, text=~paste("Sensor: ", sensor_name), mode="markers", color=~humidity, size=~humidity) %>%
|
|
|
|
|
|
|
|
layout(
|
|
|
|
|
|
|
|
plot_bgcolor='#e5ecf6',
|
|
|
|
|
|
|
|
xaxis = list( matches='x',
|
|
|
|
|
|
|
|
zerolinecolor = '#ffff',
|
|
|
|
|
|
|
|
zerolinewidth = 2,
|
|
|
|
|
|
|
|
gridcolor = 'ffff',
|
|
|
|
|
|
|
|
range = list( min(sensor_readings$timestamp),
|
|
|
|
|
|
|
|
max(sensor_readings$timestamp))
|
|
|
|
|
|
|
|
),
|
|
|
|
|
|
|
|
yaxis = list(
|
|
|
|
|
|
|
|
zerolinecolor = '#ffff',
|
|
|
|
|
|
|
|
zerolinewidth = 2,
|
|
|
|
|
|
|
|
gridcolor = 'ffff')
|
|
|
|
|
|
|
|
)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
htmlwidgets::saveWidget(as_widget(p), file="index.html")
|
|
|
|
htmlwidgets::saveWidget(as_widget(p), file="index.html")
|
|
|
|
|
|
|
|
|
|
|
|
@ -280,7 +268,7 @@ An example of a Plotly app is included in the installation. Here, we add the RP
|
|
|
|
The output should look at below:
|
|
|
|
The output should look at below:
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
.. image:: images/R-Sensor.png
|
|
|
|
.. image:: images/rplotly-postgresql.png
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
R Standard Plot (PNG)
|
|
|
|
R Standard Plot (PNG)
|
|
|
|
|

 PostGIS
PostGIS Mobile
Mobile QGIS
QGIS MapBender
MapBender GeoServer
GeoServer GeoNode
GeoNode GeoNetwork
GeoNetwork Novella
Novella Solutions
Solutions